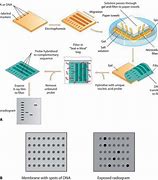
Dot blot is a technique for detecting, analyzing, and identifying proteins. This technique is similar to western blot, but protein samples are not separated using electrophoresis; instead, proteins are spotted through circular templates directly onto the membrane or paper substrate.
The concentration of proteins in crude preparations (such as culture supernatant) can be estimated semi-quantitatively using the dot blot method if you have both purified protein and the specific antibody against it. In this case, compare the signal from the unknown sample to that of the standard and estimate the concentration.
Dot blot is a technique for detecting, analyzing, and identifying proteins. This technique is similar to western blot, but protein samples are not separated using electrophoresis; instead, proteins are spotted through circular templates directly onto the membrane or paper substrate.
The concentration of proteins in crude preparations (such as culture supernatant) can be estimated semi-quantitatively using the dot blot method if you have both purified protein and the specific antibody against it. In this case, compare the signal from the unknown sample to that of the standard and estimate the concentration.
From Wikipedia, the free encyclopedia
Technique for protein detection
A dot blot (or slot blot) is a technique in molecular biology used to detect proteins. It represents a simplification of the western blot method, with the exception that the proteins to be detected are not first separated by electrophoresis. Instead, the sample is applied directly on a membrane in a single spot, and the blotting procedure is performed.
The technique offers significant savings in time, as chromatography or gel electrophoresis, and the complex blotting procedures for the gel are not required. However, it offers no information on the size of the target protein.[1]
Performing a dot blot is similar in idea to performing a western blot, with the advantage of faster speed and lower cost.[2] Dot blots are also performed to screen the binding capabilities of an antibody.[3]
A general dot blot protocol involves spotting 1–2 microliters of a samples onto a nitrocellulose or PVDF membrane and letting it air dry. Samples can be in the form of tissue culture supernatants, blood serum, cell extracts, or other preparations.[4] After the protein samples are spotted onto the membrane, the membrane is placed in a plastic container and sequentially incubated in blocking buffer, antibody solutions, or rinsing buffer on shaker. After antibody binding, the membrane is incubated with a chemiluminescent substrate and imaged.
Vacuum-assisted dot blot apparatus has been used to facilitate the rinsing and incubating process by using vacuum to extract the solution from underneath the membrane, which is assembled in between several layers of plates to ensure good seal between sample wells, hold waste solution, and deliver suction force. For chemiluminescence signal detection, apparatus need to be disassembled and the membrane need to be taken out and wrapped in a transparent plastic film.
This chapter describes the dot blot technique which is similar to the western blot one, but it is more rapid because the protein lysate is directly spotted on the membrane by means of vacuum. This strategy is useful for a qualitative analysis, e.g., a rapid screening of a large number of samples or as a quantitative technique. It is especially useful for testing the suitability of experimental design parameters (such as the optimization of primary antibody concentration).
Dot blot is a technique for detecting, analyzing, and identifying proteins. This technique is similar to western blot, but protein samples are not separated using electrophoresis; instead, proteins are spotted through circular templates directly onto the membrane or paper substrate.
The concentration of proteins in crude preparations (such as culture supernatant) can be estimated semi-quantitatively using the dot blot method if you have both purified protein and the specific antibody against it. In this case, compare the signal from the unknown sample to that of the standard and estimate the concentration.
0%0% menganggap dokumen ini bermanfaat, Tandai dokumen ini sebagai bermanfaat
0%0% menganggap dokumen ini tidak bermanfaat, Tandai dokumen ini sebagai tidak bermanfaat
%PDF-1.4 %âãÏÓ 33 0 obj <> endobj 38 0 obj <>/Filter/FlateDecode/ID[<25F54701901E2249B5B87E4EBA5C4C9C>]/Index[33 12]/Info 32 0 R/Length 47/Prev 8751/Root 34 0 R/Size 45/Type/XRef/W[1 2 1]>>stream hŞbbd``b` Œ ∫$~¹Œ€,Ftâ?ã¼? ~Vİ endstream endobj startxref 0 %%EOF 44 0 obj <>stream hŞb```f``:Ä Ş¨€ˆY8�Ř¡˜�á7«•X–‰�A2ª£B‹M�ĞLGázY´_CDÕ xj; endstream endobj 34 0 obj <>/Metadata 3 0 R/PageLabels 29 0 R/PageLayout/OneColumn/Pages 31 0 R/PieceInfo<>>>/StructTreeRoot 7 0 R/Type/Catalog>> endobj 35 0 obj <>/Font<>/ProcSet[/PDF/Text]>>/Rotate 0/StructParents 0/Type/Page>> endobj 36 0 obj <>stream hޤ’mOÛ0ǿʽÜ^d~v !µ@aÒÚ!RÁ$į5¥ûöİÙ?–�Aº‘ M)p ’&À$‚ÃÑú} ÷*o´3Æ@P6è–ÙğLÑôy¥ÑÙ“=Ϭ²U«V?>vgÍL—’” µºĞf±´ …D§ºU"J$åjÑ ãhT•v8¬�n#!ˆ��bŒ�`*ï‚:R…ÉŸ¿MM¡˜èG¸ª U~ÚDÅ aÿ2‹†U>Oƒ�ÙZÛÙMªºPyغi3Š1F?ÊÍlP.r eV×@˜lËó¾>åÚ¬lU£?]%‘à2Ô9T�ö>»ÂŸ•³jnʺ1å lÌz=2ucO–ªFßEñß‚á ıR�qO•=üµ>¡iı CfëôÜÕs»ln©ğǶ7ÆØhRÆnŒ)|@ãœØ£qfáşV¹ör9ÜÈ�|Ë �÷ãæŸÈğÈ$ô0bö†…¾ƒ%!{Áâ1èQğª·[¯ğY}wë‘ÙD§¿SÒí´ÿ:cH· Ò¢áãmÎş¶>“ï½wöfU%éÚÏwŸ]oó‘¿Ò>‰/œ�¥CşE€ J)q endstream endobj 37 0 obj <>stream H‰´VËn¤8İów £àğ¦jÔjiÒi�z4-E Ò,R³0à N¦1tMåëûÚ¼Šylf•+…�ϽçÜc.okZÁ§O—ß¿|»>¾ºş†÷†ŒË?oq-�«Ä¸L\Hö†CÇ ÉÀ—܉×QŸÏø-iÀõğÓQ±C6Äá–l7�É“a^‹Ö¾*Eu#Z‘‰òw+y0¾&Æ×ïH`ܓЃOb\âĞ0co\.H»#é%GEÏQÄfn�p³¤§§^ŞL¨ŞÕíQÏ* "â;ï@ùƒò=�P7ØÆ+ÈY˲–‹ :É«{h9ö,ÅÆ©ÓtûıY _\`í¹qgNí.Aò'kcò’6Ğ 80Ù²Æú7ù«Çq'×Õ8. Üpªô¡b�ÄÚ‚"Ñ ™e»f[ˆ\‚†³g¼qu ÛI„`£�¡Â4iYŠìE£ëâ9«Z¾çÕÓ*Ç?Z%{¬CÜ#Gr¤‰nˆã DMİ8½ƒWXÖHàZLÍziÛ ñ7c‰}_—‰RÜ!çû=k†êôæpnVÜW‘À9©îÎÜ7BAAÛĞœ«šh9ô¼ï% ¶EÅtLθgb†d»�D€ô¦ ÕbÔƒU$U‡Õ%“ƒWf%\�dP'Y‰îjD]ì—nG�VmPš YÓäKuûe<ˆàÍn©Ñ*,Q¡Ï†~FæSÚĞ ù)i‹cÚğœ?ãC�6Â�~½ ö ~B�¢WR‘Ñ”!s¦mÙ�iÿèÔ¯-öç'ƒ'Fe×è_Ñ_2<)ešU"çLyEP"iì�¦±2ÆÑê¯åŒ±ç“0z'âõ\À²¤a†!ñŞ™àC!Eş2¯A…ë….]¯–‡³„�‚-‰|ÔˆÀß4e%T]–±²ìJ!Ù<`j‘Z®©l;³÷-…š½�"‹Y¯2^î,•eCx§q`e§cdßP JX•“\ö€4ÇL!ñ²Lg|ê«J�À ¯<+2w^Z�YÂ8ïŒfÅt.� >ƒ•ÃæIĞ…Û�®üú \>¡gepãÌ:ùí”çÜøñÎÌ�Ş?ÖÖ,^Íñ‡7ëRŠdWğU”ce�ªñRŒ=ÃDv¡]ó‘óû>mÆ_s<ÿ�=†Üx0"��Ø™(�Zಠ�+ÁŒnB,L'%í g�u£ï2ÚaŠ‚©€9I¼™¡72Ü™9Ç1yÖªßYºÓmãê{ÂõI4Ç›§»ƒIÖB#Ä&¹ÎM†™ŒYsÖ5{ؽæ5oÔ°¿tµ†lĞP”,V¼3Šw ²PìÓN]PØ]şÃà€)±�3:½�íç—LNzµpı)}{ËIZÉ2Qå½(«/q/m/˜MËGZrÜŒ7�¬ñ•‚b:°êù8% ŞCw¯�ñ6^8ÙnÒ~g�ªºbÕ�Æ„Û!Efå—Î_Æ…Ş°&`0 ávğşÿp9…ñ\Z41(ÎõÚªãCS-úĞUßz²K%¾¦à£ª*°ôÔv8Y¥ºÕt)úuà'—V¨Ø¹fZ¾—Q'삉Gv·¨5³† Q¹ó¼®Sö¤ßx/òHOóø2XCsìoŞ3õL‰¥”êm«<Ï5{À{û6pé‚s¼QûFp�·ë/ ¡Ÿ‰ endstream endobj 1 0 obj <>stream hŞ„TÛ�Ú0ı•ù;±¤—¢"vYT}@<ğÒˆ#¯i—¿ïŒ�P`µô�L<çx.g&¤AYmàI<®Ràz�Ğ)ÉK@E‚LÑ(PšTŒ&¥0DR¤ Œ+2ƒ�µ#1´ESaL!1—lƒPh3äRRL„γ��Ø‹`Ʀ¹Õ•[X±È[ÇD¸±>g3Sê—üH eq>j6wö´ñ¼™1®Ó¡˜K, ”‡'¬ÙŒ"³bSH}°9›ëM¸?9Ş—éCïXsìçG6,v'«Ù·j[§Ù„CS–æ�Ş~ÇrlYT{6?æº�»"Ùü Ù øÍFrôüW�Å×á&Æò’MÙëÉÜP°Íb¯ÍÉÕØiı¾±ÅñrÄÔ×�Åk}²#z¡Ç°öêÚ3¾¨ÕÖ×Rw:KÜ ¯’øŸ‚´ŞxW¨U—æL’�IDRXÔ3ŞA,k±/ ÃÄ@ í3kZ£ñC4yˆÊ‡¨¢òÔp—vİw)ñ¢O¼09OÆ�aS•ƒ¨±WöœŸëqmt÷ÍiëışØÓoÆêpv¹uW÷h(ݲØUñç+ø§-\Qí^ÌV³g»X×]ğºAúCa‹A#lÔ@¢�fÍWæîø9Å ¥g¶ç ÁR$79‘–¯Ë0À.Ò)E‹ã•ßÓX¶T’¬®¤‘mÑâŸ:Óå8 Âz¥ÙìïåhzP_õPoAò¶óK Rp 7Ææ®0Uß”ÆúJëßê�àÿTî·“ßw�ûñW€ ÙÉn3 endstream endobj 2 0 obj <>stream hŞ2²T0P06P0´T°±Ñ÷+Í-�ó ‚bíì€BÁú.vv £GÅ endstream endobj 3 0 obj <>stream Acrobat Distiller 7.0 (Windows) Sanquin D:20100215075158 Acrobat PDFMaker 7.0 for Word 2012-11-05T15:55:09+01:00 2010-08-16T14:38:56+02:00 2012-11-05T15:55:09+01:00 uuid:98084617-47c1-489d-9468-58f3653dfb27 uuid:509c0cb0-8a3b-4363-8a2a-e7013a6c4594 2 application/pdf Dot-Blot protocol: buulj endstream endobj 4 0 obj <>stream hŞ26T0P°±ÑwÎ/Í+Q0Ô÷ÎL)�66 Åê‡T¤ê$¦§ÛÙ çoŞ endstream endobj 5 0 obj <>stream hŞDÎAk1à¿’›Äf�nEëÒ› Xğâ%n"�¦'”şûjñúx|ï�� ±X¨UáO¤æXJ:KµÆ¯«Ë?ÍÎå[‰¹GÌ�ãĞts ÕS=·v:3
Dot blot is a technique for detecting, analyzing, and identifying proteins. This technique is similar to western blot, but protein samples are not separated using electrophoresis; instead, proteins are spotted through circular templates directly onto the membrane or paper substrate.
The concentration of proteins in crude preparations (such as culture supernatant) can be estimated semi-quantitatively using the dot blot method if you have both purified protein and the specific antibody against it. In this case, compare the signal from the unknown sample to that of the standard and estimate the concentration.
Dot blot is a technique for detecting, analyzing, and identifying proteins. This technique is similar to western blot, but protein samples are not separated using electrophoresis; instead, proteins are spotted through circular templates directly onto the membrane or paper substrate.
The concentration of proteins in crude preparations (such as culture supernatant) can be estimated semi-quantitatively using the dot blot method if you have both purified protein and the specific antibody against it. In this case, compare the signal from the unknown sample to that of the standard and estimate the concentration.























